Two years ago, Chapman University doctoral student Dony Ang was using ChatGPT to write an email when the idea came to him: he could use it to design medication.
“Instead of inputting the text prompts, what you can enter into the chatbot is the sequence of the protein causing the disease,” says Ang, who is completing his Ph.D. in computational data science and molecular biology while working as a data scientist for a health care company.
Synthetic drug creation can target proteins to halt the spread of cancer and other diseases. But current methods can be labor-, time- and cost-intensive.

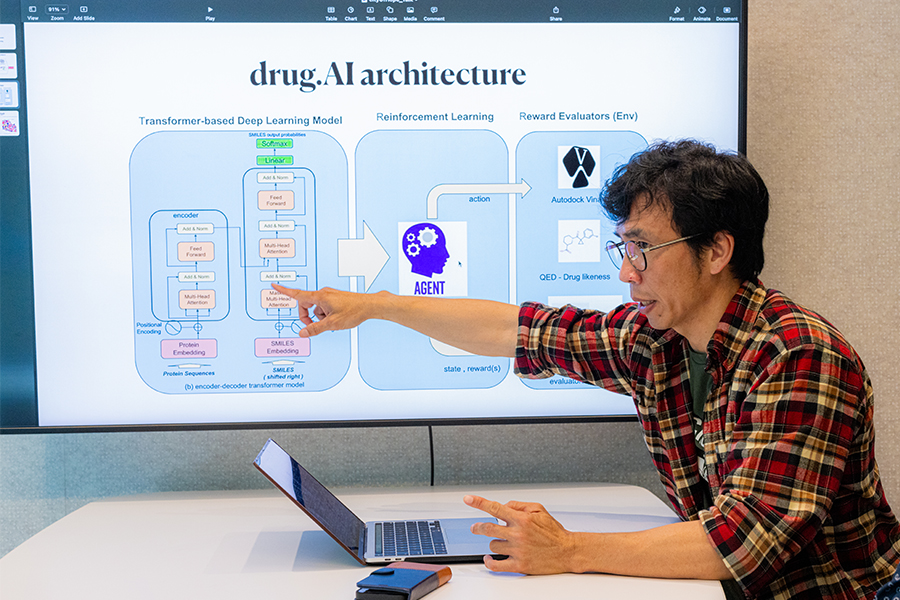
In Ang’s new model, which he calls drug.AI and details in a paper published in January with two Chapman professors, the building blocks of ChatGPT and a reinforcement-learning algorithm called Monte Carlo Tree Search (MCTS) combine to quickly generate drug compounds that can target the protein causing a disease.
One of Ang’s co-authors, Assistant Professor Hagop Atamian, likens it to creating a key to fit a lock.
“Instead of spending weeks to screen millions and millions of available molecules and seeing which ones fit, like going to a warehouse with millions of keys and looking for one that fits, the keymaker knows by just looking at the lock that they need to create this type of key,” he says.
Ang had already been working on finding inhibitors for COVID-19 proteins when he got the idea to use an approach similar to ChatGPT to design drugs. The COVID research took around a year.
“It took a long time to screen thousands of molecules to find ones that bind to our target — it’s not plug and play like drug.AI,” Ang says. “It’s super laborious and super time consuming and super hard, especially when it comes to cancer.”
In drug.AI, a disease’s target protein sequence is entered into a prompt and potential inhibiting molecules are generated within hours or even minutes. Drug.AI refines drug compounds for essential qualities like safety for human consumption, likelihood that it can be synthesized by a chemist and binding affinity, or how tightly drug molecules bind to the disease protein.
According to Ang’s and Atamian’s paper, the molecules generated by drug.AI were of similar quality to those from two other common methods, and in some cases, better. They found that drug.AI’s candidate drugs had a validity rate of 100%.
Drug.AI’s candidate drugs were also measured for drug-likeness, or the similarity of a compound’s properties to those of oral drugs, and candidate drugs were 42% to 75% higher than other models. Additionally, all drug.AI-generated molecules showed strong binding affinities to targets, comparable to those identified through traditional virtual screenings.
Ang and Atamian also looked at how drug.AI’s results for a specific disease compared to known drugs for that disease. They compared drug-likeness and binding affinity between the natural molecules and drug.AI’s, and found similar measurements in both — but drug.AI was able to identify these in a much quicker and less costly way.
Before a method like drug.AI, it was time-consuming for databases to screen drug candidates for synthesis and eventual FDA approval.
“No matter how fast they made it, it still needed something like a supercomputer,” Atamian says.
“Now, we are able to do this on our laptops in an hour.”
Previous drug candidates were also limited to what was already in existence.
In drug.AI, Ang teaches the program to create a drug for a specific disease. In graphics showing how drug.AI works, there is a cavity on a disease protein that a drug fits into, or binds to.
“If it’s a protein involved in cancer proliferation, your molecule binds to it and it doesn’t let the protein do its job and the cancer doesn’t progress,” Atamian says.
The speed at which drugs can be customized to a disease — pending how long it takes a chemist to synthesize them — will be the “game changer,” Ang says.
“During the pandemic, we didn’t have much time and there was a deadly virus. And it took us time to develop drugs,” he says. “Hopefully, this can be a lifesaver.”
The researchers want to open source their method so others can use it. Atamian, an agricultural researcher, says it could be used to stop crop diseases.
Other Chapman researchers are exploring ways of using AI in drug development, including molecule vetting.




